Examples of distributions with singularities
| ... from functools import partial
from pylab import *
from mpl_toolkits.axes_grid.inset_locator import zoomed_inset_axes
from mpl_toolkits.axes_grid.inset_locator import mark_inset
from pacal import *
|
||
|
.
|
||
Using compiled interpolation routine
| ... from pacal.distr import demo_distr
|
||
|
.
|
||
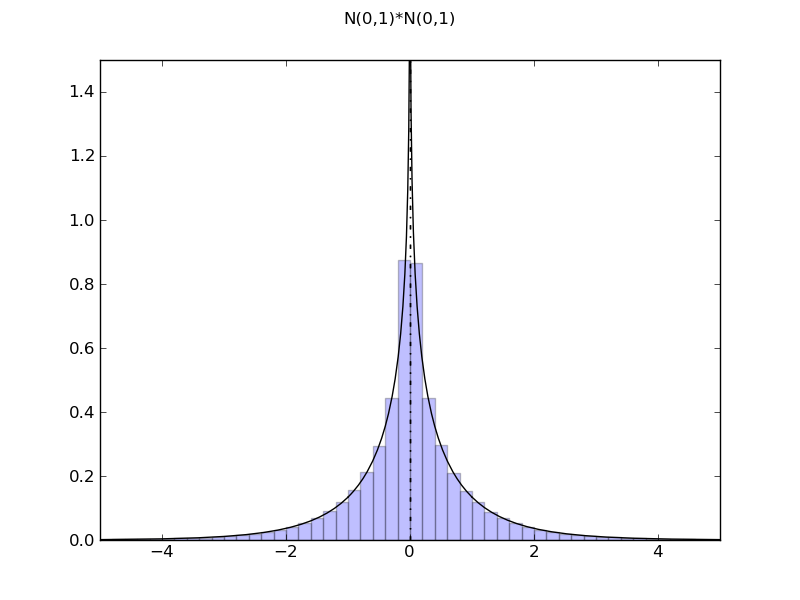
Product of two shifted normal variables
such a product always has a singularity at 0, but the further the factors' means are from zero, the 'lighter' the singularity becomes
| ... figure()
d = NormalDistr(0,1) * NormalDistr(0,1)
demo_distr(d, ymax=1.5, xmin=-5, xmax=5)
|
||
|
.
|
||
============= summary =============
N(0,1)*N(0,1)
mean = 5.55111512313e-17
std = 1.0
var = 1.0
tailexp = (-148.1820750072207, -148.1820750072207)
median = 0.0
medianad = 0.365168011835
iqrange(0.025) = 4.36389796793
range = (-inf, inf)
ci(0.05) = (-2.1819489839664534, 2.1819489839664223)
int_err = -8.881784197e-16
| ... show()
|
||
|
.
|
||
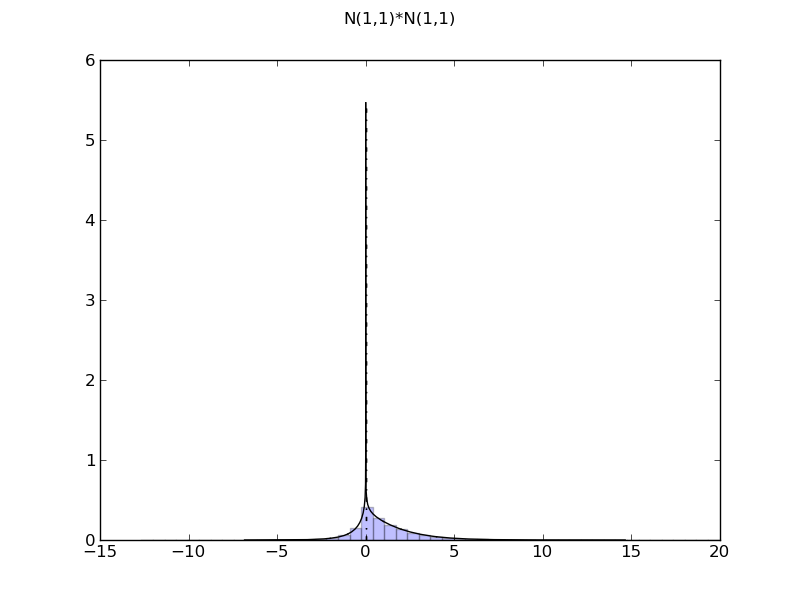
| ... figure()
d = NormalDistr(1,1) * NormalDistr(1,1)
demo_distr(d)
|
||
|
.
|
||
============= summary =============
N(1,1)*N(1,1)
mean = 1.0
std = 1.73205080757
var = 3.0
tailexp = (-148.18120012361717, -136.11316051087496)
median = 0.608550006303
medianad = 0.795218786198
iqrange(0.025) = 7.03600471957
range = (-inf, inf)
ci(0.05) = (-1.7253932992908485, 5.3106114202823536)
int_err = -6.66133814775e-16
| ... show()
|
||
|
.
|
||
| ... figure()
d = NormalDistr(2,1) * NormalDistr(2,1)
demo_distr(d)
|
||
|
.
|
||
============= summary =============
N(2,1)*N(2,1)
mean = 4.0
std = 3.0
var = 9.0
tailexp = (-148.17862539773668, -124.04524124537676)
median = 3.53736652838
medianad = 1.89598902303
iqrange(0.025) = 11.4531157325
range = (-inf, inf)
ci(0.05) = (-0.3998570580771057, 11.053258674424656)
int_err = 0.0
| ... show()
|
||
|
.
|
||

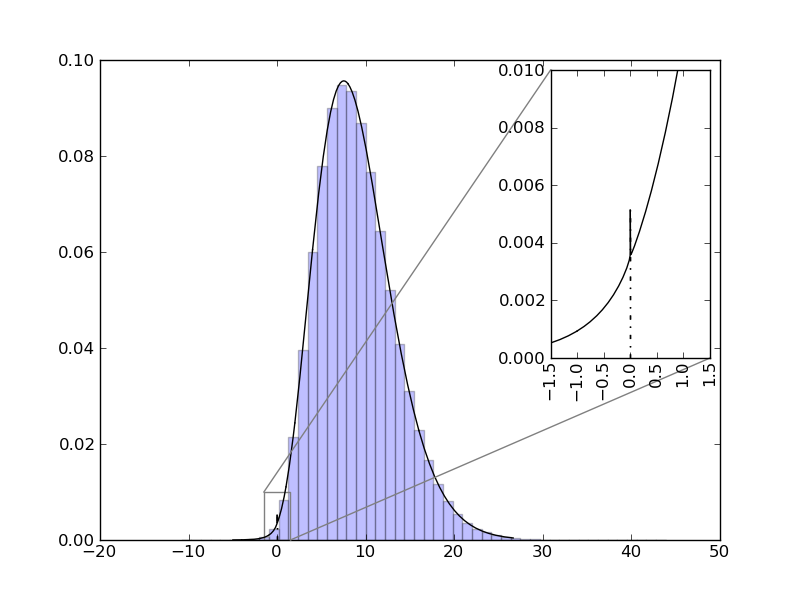
| ... figure()
d = NormalDistr(3,1) * NormalDistr(3,1)
d.plot()
d.hist()
ax=gca()
axins = zoomed_inset_axes(ax, 6, loc=1)
d.plot(xmin=-1.5, xmax=1.5)
axins.set_xlim(-1.5, 1.5)
xticks(rotation="vertical")
axins.set_ylim(0, 0.01)
mark_inset(ax, axins, loc1=2, loc2=4, fc="none", ec="0.5")
show()
|
||
|
.
|
||
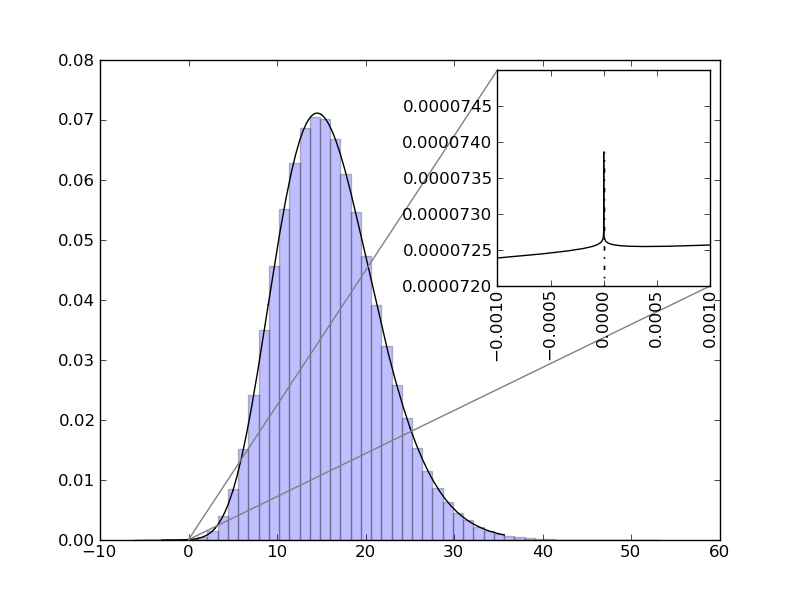
| ... figure()
d = NormalDistr(4,1) * NormalDistr(4,1)
d.plot()
d.hist()
ax=gca()
axins = zoomed_inset_axes(ax, 12000, loc=1)
d.plot(xmin=-.001, xmax=.001)
axins.set_xlim(-.001, .001)
xticks(rotation="vertical")
axins.set_ylim(0.000072, 0.000075)
mark_inset(ax, axins, loc1=2, loc2=4, fc="none", ec="0.5")
show()
|
||
|
.
|
||
| ... |
||
|
.
|
||
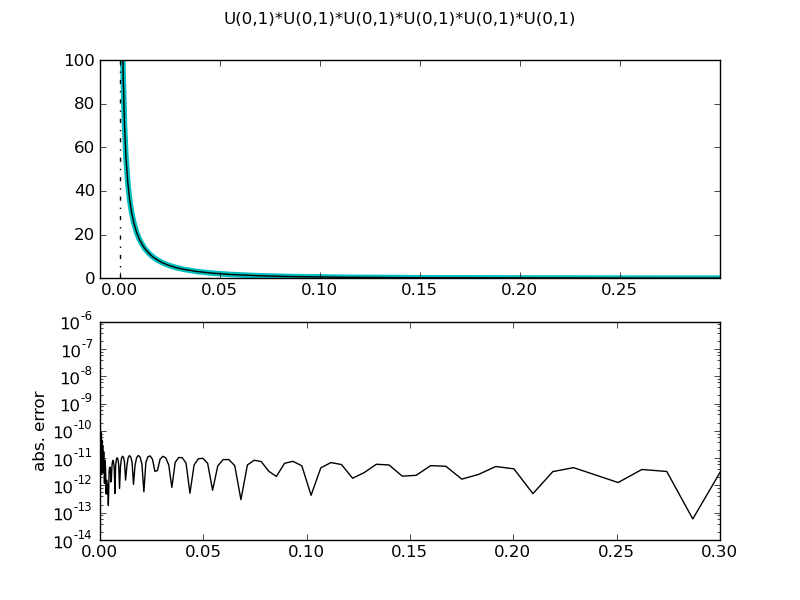
Product of six uniform distributions
| ... def prod_uni_pdf(n, x):
pdf = (-log(x)) ** (n-1)
for i in xrange(2, n):
pdf /= i
return pdf
figure()
d = UniformDistr(0,1) * UniformDistr(0,1) * UniformDistr(0,1) * UniformDistr(0,1) * UniformDistr(0,1) * UniformDistr(0,1)
demo_distr(d, ymax=100, xmin=-0.01, xmax=0.3, theoretical = partial(prod_uni_pdf, 6))
|
||
|
.
|
||
============= summary =============
U(0,1)*U(0,1)*U(0,1)*U(0,1)*U(0,1)*U(0,1)
mean = 0.015625
std = 0.0335797779546
var = 0.00112760148748
tailexp = (None, None)
median = 0.00344730956418
medianad = 0.003321699878
iqrange(0.025) = 0.110584907089
range = (0.0, 1.0)
ci(0.05) = (8.5606702109140504e-06, 0.11059346775905532)
int_err = -4.4408920985e-15
max. abs. error 7.82310962677e-07
max. rel. error 1.42505744692e-10
| ... show()
|
||
|
.
|
||
| ... |
||
|
.
|
||